GNATS of unknown function
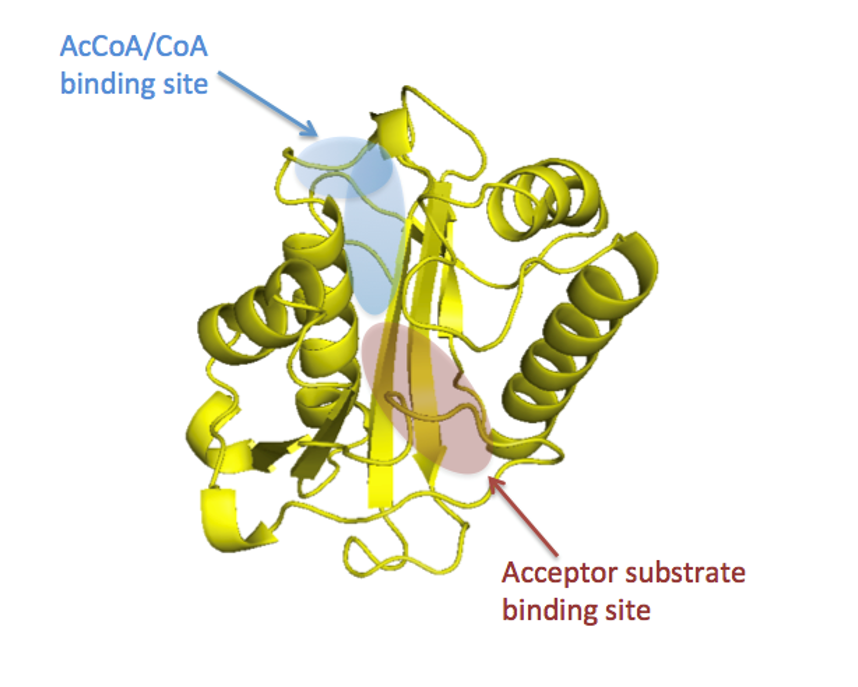
We study bacterial GNATs of unknown function and develop chemical tools to obtain crystal structures with ligands bound to the acceptor sites for improved computational predictions of protein function.
While the core structure of GNATs is conserved across members of the GNAT superfamily, the acceptor substrate binding site varies and dictates the type of substrate that becomes acylated. The more we know about the types of molecules that can bind in this site and how the amino acid composition/properties of this site contribute to specific functions, the more accurate our computational predictors of protein function will be. Since each bacterial genome encodes multiple GNATs with various functions, but not all bacteria contain the same types of GNATs, it is important to study a variety of GNATs across multiple bacteria to ensure functional potential is widely explored and accurately predicted.
We previously developed a broad-substrate screen as a tool to explore possible functions of GNATs. We continue to use this screen as a starting point for characterizing GNATs of unknown function. Once we have identified a possible substrate for a previously uncharacterized enzyme, we further kinetically and structurally characterize it to determine how the enzyme works. By exploring many different types of GNATs, we are better positioned to gain a global understanding of key mechanisms of GNAT function.