Courses with an authentic research focus
My approach to teaching biochemistry in my research laboratory and traditional classrooms (lecture and laboratory) is to create authentic research experiences whenever possible.
Chem 699, Chem 895, and Chem 897 are traditional research courses.
Highlights for non-traditional research courses Chem 443 and Chem 800 are found below.
Chem 443: Biophysical Chemistry Laboratory
Collaboration between San Francisco State University and
Argonne National Laboratory, Spring 2019
Undergraduate students in course
Ashley Alcala, Oscar Luna, Karen Nguyen, Guadalupe Ramirez, Allan Solis, Daniel Vazquez, and Michael Ward
Prior to this course 4/7 students had never performed research.
Course Information
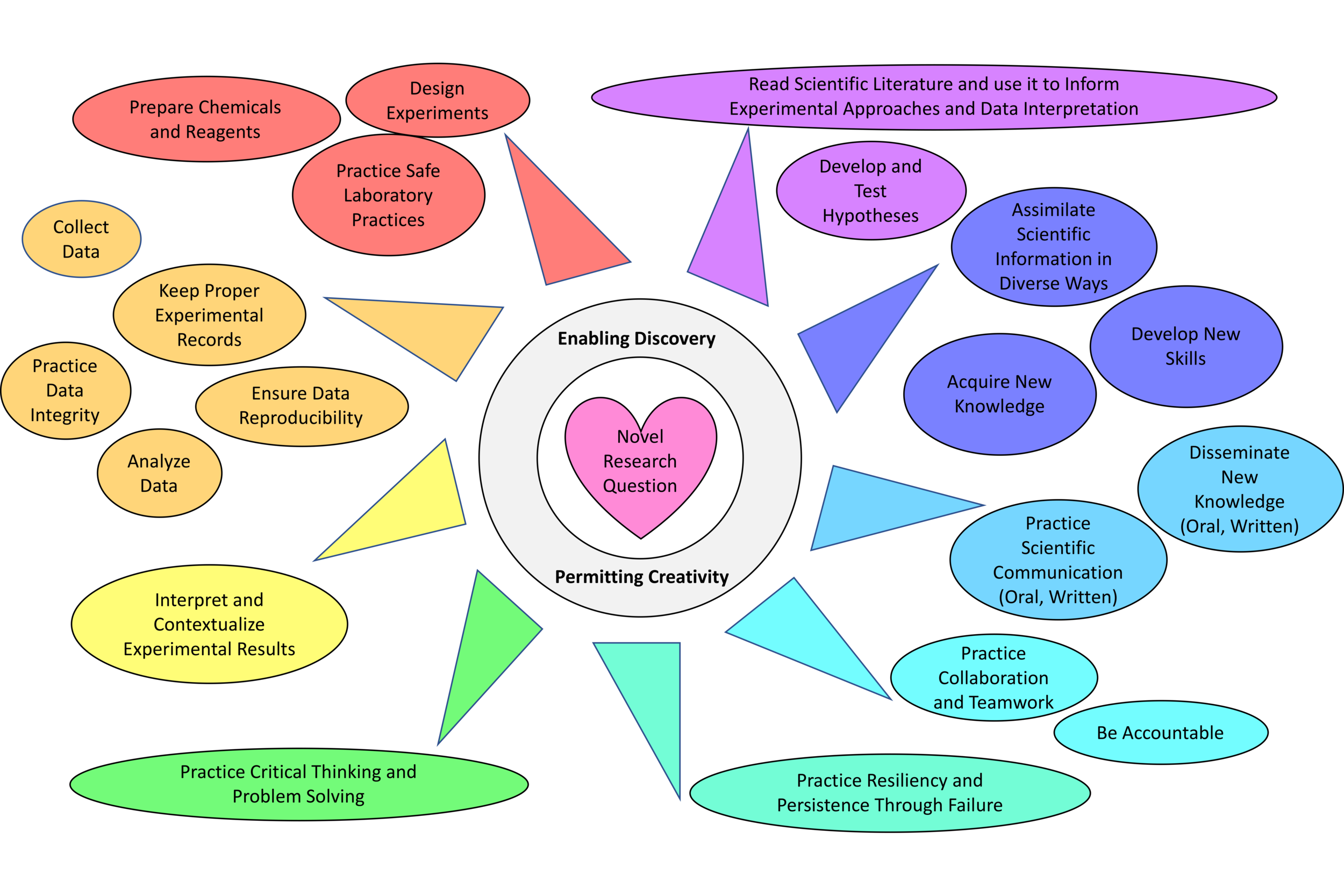
This course was designed to engage students in an authentic research experience where the instructor, collaborators, and students did not know the outcome of the experiments. The rationale for having students collaborate with scientists at Argonne National Laboratory was to hold them accountable, practice communicating science with others outside their institution, and to learn new knowledge from diverse scientists in a different environment than SFSU. Students also learned to work as a team to answer a novel biochemical/biophysical research question and present their results at the CoSE Showcase. This course offered an opportunity to engage some students in research who had never performed research prior to this class and enable them to develop skills needed for their next steps in their careers after college.
The heart of the course is centered on a novel biochemical/biophysical research question. The role of the instructor is to guide students as they pursue new knowledge, enable independent and team discovery, and permit creativity in the way experiments are performed and results are presented.
Scientific manuscript published as a result of student work in course
SFSU Student co-authors are denoted with #
Alcala A.#, Ramirez G.#, Solis A.#, Kim Y., Tan K., Luna O.#, Nguyen K.#, Vazquez D.#, Ward M.#, Zhou M., Mulligan R., Maltseva N., and Kuhn M.L. (2020) Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species. Protein Sci. 29(3), 695-710. PMID: 31762145. doi: 10.1002/pro.3793
Protein crystals obtained for this project by Natalia Maltseva
Student Presentations at the 2019 SFSU CoSE Showcase
Chem 800: Protein Structure Function
Collaboration between San Francisco State University and
Northwestern University Feinberg School of Medicine, Spring 2017
Featured article about the course
https://news.sfsu.edu/news-story/biochemistry-grad-students-go-above-and-beyond-publications
Graduate students in course
Jackson Baumgartner, Trevor Gokey*, Ashley Law, Jesi Lee, Jennifer Macias, Alexander Stergioulis, Ha Tran*
Undergraduate student in course
Aleksander Shornikov*
*Not pictured
Course Information
This course was designed to help students gain a basic knowledge of protein structure and function concepts and 3D structure visualization using computer software and web-based tools. This was accomplished through the lens of constructing a scientific manuscript for publication. The rationale for this particular approach to the course was two-fold: 1) students are more engaged in learning new concepts when they have ownership in the learning process, and 2) writing scientific manuscripts for publication will aid the professional development of students and improve scientific literacy.
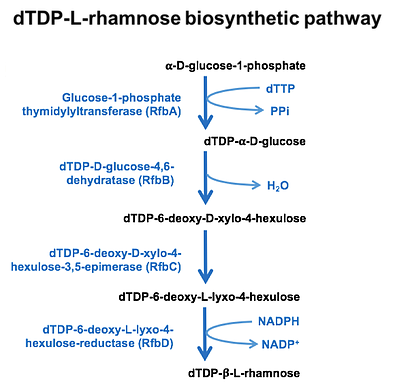
The focus of the course was to perform structural comparisons and analyses of 3D structures of enzymes belonging to the dTDP-L-rhamnose biosynthetic pathway from Bacillus anthracis. Since the proteins of this pathway catalyze diverse reactions and have different structural folds, students were exposed to a variety of protein structures. Focusing on a single metabolic pathway also encouraged collaboration between students, provided an opportunity to analyze their individual proteins in the context of a larger theme, and allowed them to think about roles of their individual proteins in regulating the overall pathway.
Scientific manuscripts published as a result of student work in course
SFSU Student co-authors are denoted with #
RfbA
Baumgartner J.#, Lee J. #, Halavaty A.S., Minasov, G., Anderson W.F., and Kuhn M.L.& (2017) Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme glucose-1-phosphate thymidylyltransferase (RfbA). Acta Cryst. F73, 621-628. PMID: 29095156. doi: 10.1107/S2053230X17015849
RfbB
Gokey T. #, Halavaty A.S., Minasov G., Anderson W.F., and Kuhn M.L.& (2018) Structure of the Bacillus anthracis dTDP-L-rhamnose biosynthetic pathway enzyme: dTDP-α-D-glucose 4,6-dehydratase, RfbB. J. Struct. Biol. 202(2), 175-181. PMID: 29331609. doi: 10.1016/j.jsb.2018.01.006
RfbC
Shornikov A.#, Tran H. #, Macias J. #, Halavaty A.S., Minasov G., Anderson W.F., and Kuhn M.L.& (2017) Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme dTDP-4-dehydrorhamnose 3,5-epimerase (RfbC). Acta Cryst. F73, 664-671. PMID: 29199987. doi: 10.1107/S2053230X17015849
RfbD
Law A.#, Stergioulis A.#, Halavaty A.S., Minasov G., Anderson W.F., and Kuhn M.L.& (2017) Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme dTDP-4-dehydrorhamnose reductases (RfbD). Acta Cryst. F73, 644-650. PMID: 29199984. doi: 10.1107/S2053230X17015746